Machine Learning to increase biotechnology-based protein production
In a research co-operation, researchers of the Paul-Ehrlich-Institut (PEI) have developed a mathematical model which allows more accurate forecasts and improved output in biotechnology-based protein synthesis in widely used host organisms. The new method offers many and varied applications in biotechnology including the development of vaccines. Scientific Reports has published an article on the results in its online version of 17 May 2019.
Biotechnology medicinal products are frequently based on tailor-made proteins produced in cell cultures or bacteria. For this purpose, the genes containing the information on the amino acid sequence of the desired proteins are transferred to bacterial or mammalian cells. However, this is often not sufficient to read the transferred genes to the desired efficiency and to form the proteins coded on them. Usually, an additional adaptation of the respective genes in the host cell is required. Among other things, this happens by adaptation of the code for the amino acids. The subsequences of three nucleobases each of the messenger RNA (mRNA), also called codon, determine the individual amino acids; the sequence of the codons determines the amino acid sequence of proteins. An exchange of these codons is necessary because different organisms, i.e. cell systems have different codon preferences for one and the same amino acid. The reason for this has scientifically not yet been fully understood. The adaptation of the codons has therefore so far been made using a heuristic approach.
How can it be better predicted which optimisation steps are suitable? In a research co-operation supported by the Adolf-Messer Foundation with researchers of the Max Planck Institute for Colloids and Interfaces, Potsdam, and the Goethe University at Frankfurt/Main, co-workers of Dr Jan-Hendrik Trösemeier and Dr Christel Kamp, Section Biostatistics of Division Microbiology of the Paul-Ehrlich-Institut studied the protein expression in the so-called codon-specific elongation model (COSEM). In this study, mathematical methods are used to simulate the dynamics of the protein synthesis (protein translation) in the appropriate cells and a codon-specific rate of protein synthesis is derived from this.
Using the data of this simulation, the researchers have found the so-called protein expression score, taking into account additional predictors for the protein output and using methods of "machine learning". This protein expression score serves to forecast the protein output and to optimise the codons of the genes, which are (heterologously) expressed in foreign cells. In various model organisms, the researchers provided proof that their simulation-based optimisation method was superior to conventional methods. Not only can the protein output be increased with this newly developed modular model, but further optimisations can also be performed, e.g. the accuracy of translation can be improved.
The algorithm is implemented in special software programs and permits the above-described user-defined optimisation of genes. The algorithm can also be used for the inverted path – de-optimisation. What is the purpose of this? Such a de-optimisation of genes can, among other things, be used for the genetic modification and attenuation of pathogens. Such an attenuation of pathogens is used in vaccine development: Live vaccines are derived from original pathogens and are genetically modified in such a way that although they produce an immune reaction in humans, they only replicate to a limited extent, and are therefore no longer able to produce a disease.
This new approach to optimising codons has brought about a patent registration.
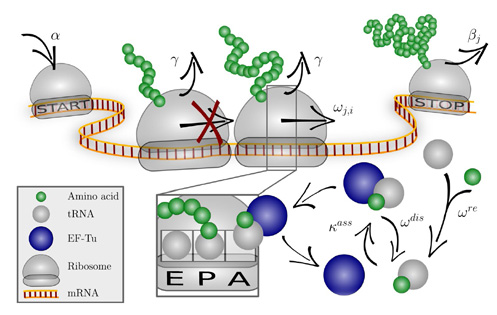 The codon-specific elongation model (COSEM) simulates protein synthesis.
Source: Scientific Reports
The codon-specific elongation model (COSEM) simulates protein synthesis.
Source: Scientific Reports
Original Publication
Trösemeier JH, Rudorf S, Loessner H, Hofner B, Reuter A, Schulenborg T, Koch I, Bekeredjian-Ding I, Lipowsky R, Kamp C (2019): Optimizing the dynamics of protein expression.
Sci Rep 9: 7511.
Text
Contact:
Paul-Ehrlich-Institut
Press Office
Telefon: +49 6103 77 1030
Email: presse@pei.de
top



